Visualize This! visualization competition
Scientific Dataset Details
This dataset is now available for downloading.
This dataset is from a coarse-grained Gromacs molecular dynamics (MD) simulation of interaction of a large protein structure with a cell’s membrane. A cell’s membrane is typically composed of two leaflets (single molecular layers), comprised in this model of pure phosphatidylcholine (POPC) lipid molecules. The protein structure is a P-glycoprotein (P-gp) embedded into the cell’s membrane. Such proteins are responsible for the translocation of a wide range of compounds across membranes.
A simple visualization with VMD (below) shows the horizontal cell membrane along with the embedded vertical protein. To further differentiate the two, we rendered the protein beads as larger spheres.
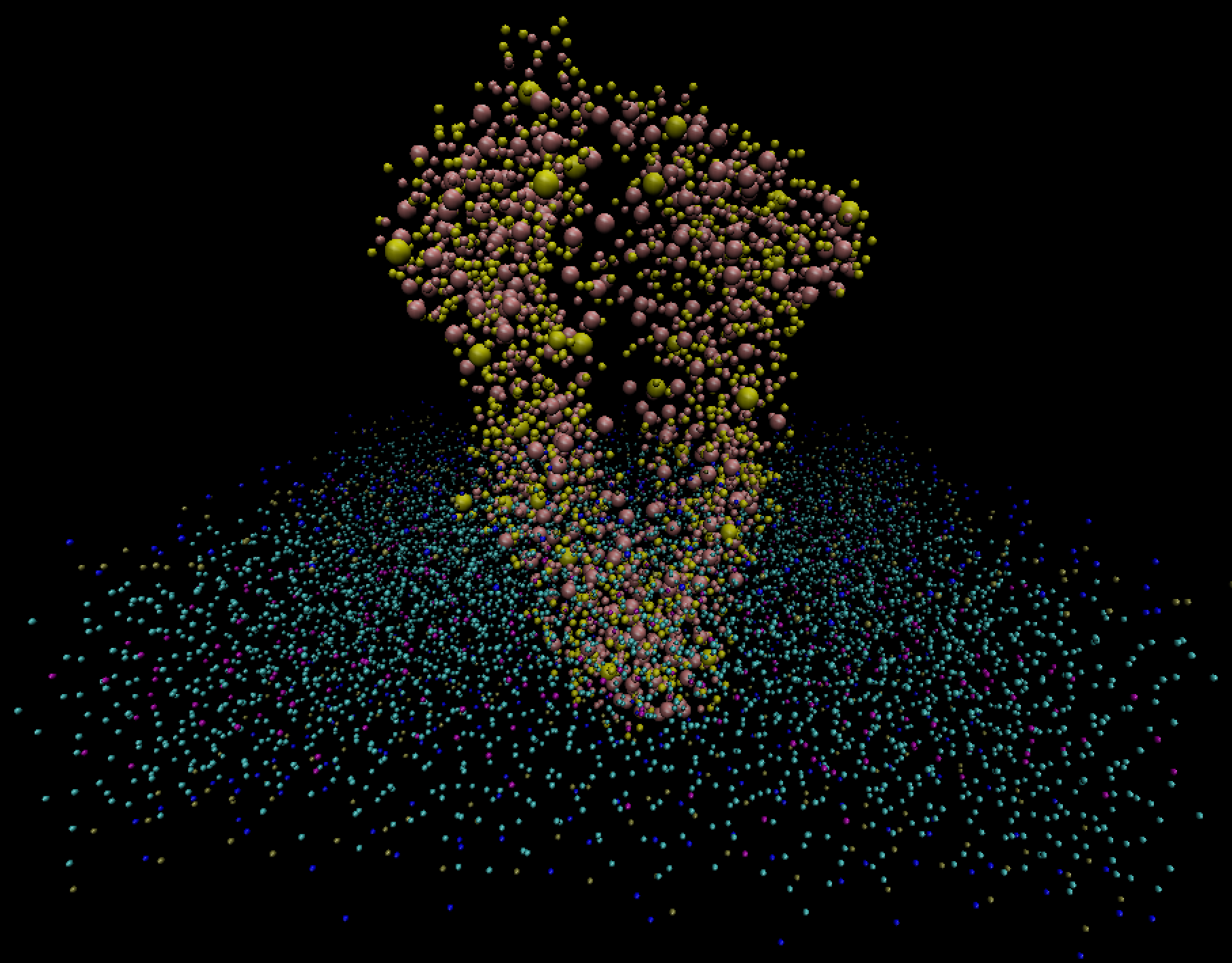
This simulation is using so-called coarse-grained MARTINI force field, in which each bead (the smallest resolution element) represents a chemical group (3-4 atoms or more), not a single atom. Groups of these beads form residues which make molecules. Typically, smaller molecules have only one residue; large molecules, like proteins or DNA, can have multiple residues.
The dataset contains 21 residue types. One of them is POPC, the lipid molecule that forms the membrane
bilayer. The rest are amino acids forming the protein, and their names are all spelled in the file
martini_v2.2_aminoacids.itp.
There are 17 bead types, abbreviated as BB, C{1..2}A, C{1..3}B, C4{A..B}, D3A, GL{1..2}, NC3, PO4, and SC{1..4} (using the brace expansion notation).
Water molecules were present in the simulation but were removed from the output to save storage space.
How to read the data
The dataset was stored in the standard Gromacs trajectory output format, so it can be easily read with any compatible software, e.g., with VMD or with MDAnalysis Python library. Let us first go through the individual files in the dataset.
Dataset format
The file sim.part0001_protein_lipid_popc.gro describes the initial setup with 9453 beads and 1756
residues in the standard GRO
format, stored as human-readable ASCII. The main body of the file lists all beads, one line per
bead. Here we describe formatting of all fields using the first bead as an example:
' 1' is the residue number (5 characters): 1 to 1756
'PRO ' is the residue name (5 characters): 21 unique names
ALA=alanine ARG=arginine ASN=asparagine ASP=aspartate
CYS=cysteine GLN=glutamine GLU=glutamate GLY=glycine
HIS=histidine ILE=isoleucine LEU=leucine LYS=lysine
MET=methionine PHE=phenylalanine POPC=ourLipid PRO=proline
SER=serine THR=threonine TRP=tryptophan TYR=tyrosine
VAL=valine
' BB' is the bead name (5 characters): 17 unique elements
BB=backbone C{1..2}A C{1..3}B C4{A..B} D3A GL{1..2}
NC3=choline PO4=phosphate SC{1..4}
' 1' is the bead number (5 characters): 1 to 9453
' 6.692' is x-position in nm (8 characters)
' 8.569' is y-position in nm (8 characters)
' 8.462' is z-position in nm (8 characters)
' 0.0038' is x-velocity in nm/ps = km/s (8 characters)
' -0.0789' is y-velocity in nm/ps = km/s (8 characters)
' -0.1406' is z-velocity in nm/ps = km/s (8 characters)
The files 5us_traj_popc_protein_lipid_popc.xtc and 15us_traj_protein_lipid_popc.xtc store the
trajectory in the standard
XTC format during the first 5 microseconds (2502 frames) and the last 15 microseconds (8502 frames)
of the simulation, respectively. These files store data as binary. Note that this file format is lossy in
the sense that saved positions are approximate.
The file system.top and the four included martini_v2.*itp files store a high-level description of the
system, the topology data (chemical bonds), the description of interactions (force field), and
bibliography. All of these data are auxiliary and are not created by the simulation: they are only
provided for reference.
The file pgp_m_A+pgp_m_B.itp contains information about the protein structure and is optional in the
analysis.
Reading data with VMD
Loading this dataset into VMD is straightforward. First you load the molecule (the Gromacs GRO file), then into this molecule you load the trajectory (XTC) file and wait for all frames to finish loading. On Unix-like systems (Linux, Mac) you can automate reading with a single command:
$ vmd sim.part0001_protein_lipid_popc.gro 5us_traj_popc_protein_lipid_popc.xtc
The visualization at the top of this page was produced with Graphics → Representations →
Drawing Method = Beads. You can easily navigate the dataset by showing different beads and residues,
using VMD
Selection Language in Graphics → Representations → Selected Atoms field:
all # show all beads
resname POPC # the entire POPC lipid bilayer
resid 1181 # one of the "travelling" lipid molecules
not resname POPC # P-glycoprotein made of all amino acids
name GL1 # one of the "beads" in POPC to see the two sheets
In the visualization at the top of this page we displayed the selection not resname POPC as larger
spheres and resname POPC as smaller spheres.
VMD is a very popular tool for visualizing such molecular structures, and we would like participants to go beyond the basics to produce more complex and informative visualizations. In the next section we show how you can process these data with MDAnalysis in Python. You can filter anything using your Python code and then display these 3D data in ParaView or VisIt.
Reading data with MDAnalysis
After you install MDAnalysis (pip install
--upgrade MDAnalysis ) and download the data, you can analyze it in Python:
import MDAnalysis as mda
u = mda.Universe('sim.part0001_protein_lipid_popc.gro', '5us_traj_popc_protein_lipid_popc.xtc')
print(u.trajectory) # 2501 frames of 9453 beads
for timestep in u.trajectory:
print(timestep)
for a in u.atoms:
print(a)
u.trajectory[0] # load the first frame
u.atoms.positions # print the positions of all beads
u.atoms.n_atoms # 9453 beads
u.atoms.n_residues # 1756 residues
bb = u.select_atoms("name BB") # group with 1180 beads
print(bb)
bb.positions # positions of only BB atoms
# MDAnalysis can select protein by identifying its amino acid components
protein = u.select_atoms('protein') # 2541 beads
protein.atoms.n_residues # 1180 residues
for p in protein:
print(p)
# can repeat the same analysis for any other frame, e.g.
u.trajectory[-1] # load the last frame
Research questions
This simulation shows that the P-gp molecule acts as a lipid transporter protein, flipping membrane’s lipids from the inner to the outer leaflet. We would like you to visualize this process, finding those lipids that bind to the protein.
For example, it would be nice to see specific interactions between lipids and protein, the frequency of the interaction and for how long such interaction is occurring. You can do this by considering the PO4 bead of the lipids and its interactions with SC{1..4} beads in the cavity of the protein.
It could be interesting to show any other correlations or unusual features in the data. You could look at the differences in the system between the beginning and the end of the simulation, when equilibrium is assumed to be achieved. You could also visualize the differences between lipids in close proximity to the protein and at the periphery. Another possibility is to build a view from a certain atom’s perspective, given that it provides some interesting insight.
You can use any open-source software to visualize the data, including VMD, PyMol, ParaView, VisIt, but your visualization should be sufficiently innovative (and reproducible!).
For more visualization examples, please check the simulation paper (E. Barreto-Ojeda et al. 2017) behind this dataset. The dataset corresponds to what is called system S1 in Table 1. Figure 1 shows two bead trajectories in the POPC bilayer, while Figure 7 plots lipid-uptake pathways through the cell membrane.